Generate valid lists of bonds for a given RNA sequence using logic to avoid contradictory bond assignments
In this example, we will generate typical sets of bonds for a small RNA sequence. First, will build a quick way to check if a bond follows the usual complementary pairing rules.
Obtain all the typical bonding pairs for RNA.
In[16]:=
bondingPairs=List@@@Normal[Entity["BioSequenceType","RNA"]["ComplementLetterRules"]]
Out[16]=
{{U,A},{V,B},{G,C},{H,D},{C,G},{D,H},{M,K},{K,M},{N,N},{Y,R},{S,S},{A,U},{B,V},{W,W},{R,Y}}
Use an Association to store these pairs so we can quickly determine if we have a complement pair.
In[17]:=
bondingPairAssoc=<|Rule[#,True]&/@bondingPairs|>
Out[17]=
{U,A}True,{V,B}True,{G,C}True,{H,D}True,{C,G}True,{D,H}True,{M,K}True,{K,M}True,{N,N}True,{Y,R}True,{S,S}True,{A,U}True,{B,V}True,{W,W}True,{R,Y}True
With this, we can make a function to build a matrix of possible complement pairings.
In[18]:=
makeFoldingMatrix[seq_BioSequence]:=With[{characters=Characters[seq["SequenceString"]],matrixLength=seq["SequenceLength"]},SparseArray[Table[Boole[KeyExistsQ[bondingPairAssoc,{characters[[i]],characters[[j]]}]],{i,1,matrixLength},{j,1,matrixLength}]]]
Let us see the folding matrix of a test sequence.
In[19]:=
seq=BioSequence["RNA","CGAUGCGAGUGG",{}];
In[20]:=
(foldingMatrix=makeFoldingMatrix[seq])//MatrixForm
Out[20]//MatrixForm=
0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
With the folding matrix, we want to find all combinations of those bonds into bond lists, but we cannot include two or more bonds sharing the same coordinate.
Now, we can extract the individual bonds from the folding matrix, only considering those bonds that aren't too close together.
In[21]:=
bondPositions=With[{matrixLength=Length[foldingMatrix]},Flatten[Map[Function[{index},{index,#}&/@Select[Range[index+4,matrixLength],1===foldingMatrix[[index]][[#]]&]],Range[matrixLength]],1]]
Out[21]=
{{1,5},{1,7},{1,9},{1,11},{1,12},{2,6},{3,10},{4,8},{6,11},{6,12}}
The way we will make compatible bond lists is by making a logical expression of which bond position pairs can be included with which other bond position pairs, and then by finding satisfying assignments for that logical expression. We will have a variable indicating whether a given bond position is included or not.
We say that if two sets of bond positions share a point, there is a conflict.
In[22]:=
conflictingPairQ[p1_,p2_]:=Intersection[p1,p2]=!={}
If a bond position pair is included, it implies that all of conflicting bond position pairs are not, and vice-versa.
In[23]:=
buildConflictImplicationsForBond[bond_,var_,remainingBonds_List,remainingVars_List]:=MapThread[If[conflictingPairQ[bond,#1],And[Implies[var,Not[#2]],Implies[#2,Not[var]]],Nothing]&,{remainingBonds,remainingVars}]
Find all of these implications for all bonds recursively.
In[24]:=
buildConflictImplications[{bond_,remainingBonds__},{var_,remainingVars__}]:=Flatten[{buildConflictImplicationsForBond[bond,var,{remainingBonds},{remainingVars}],buildConflictImplications[{remainingBonds},{remainingVars}]},1]
With a single bond and variable, there are no conflicts remaining as there is nothing to conflict with.
In[25]:=
buildConflictImplications[{_},{_}]:={}
A proper bond list includes at least one bond and respects all conflicts.
In[26]:=
buildBooleanExpressionForBondSelection[bondList_,vars_]:=And[Apply[Or,vars],Sequence@@(buildConflictImplications[bondList,vars])]
Given all this, produce the logical expression which expresses these constraints.
In[27]:=
buildBooleanExpressionForBondSelection[bondPositions,Table[Unique["bonVar"],{Length[bondPositions]}]]
Out[27]=
(bonVar11||bonVar12||bonVar13||bonVar14||bonVar15||bonVar16||bonVar17||bonVar18||bonVar19||bonVar20)&&(bonVar11!bonVar12)&&(bonVar12!bonVar11)&&(bonVar11!bonVar13)&&(bonVar13!bonVar11)&&(bonVar11!bonVar14)&&(bonVar14!bonVar11)&&(bonVar11!bonVar15)&&(bonVar15!bonVar11)&&(bonVar12!bonVar13)&&(bonVar13!bonVar12)&&(bonVar12!bonVar14)&&(bonVar14!bonVar12)&&(bonVar12!bonVar15)&&(bonVar15!bonVar12)&&(bonVar13!bonVar14)&&(bonVar14!bonVar13)&&(bonVar13!bonVar15)&&(bonVar15!bonVar13)&&(bonVar14!bonVar15)&&(bonVar15!bonVar14)&&(bonVar14!bonVar19)&&(bonVar19!bonVar14)&&(bonVar15!bonVar20)&&(bonVar20!bonVar15)&&(bonVar16!bonVar19)&&(bonVar19!bonVar16)&&(bonVar16!bonVar20)&&(bonVar20!bonVar16)&&(bonVar19!bonVar20)&&(bonVar20!bonVar19)
We can now find valid combinations of bonds.
In[28]:=
findSatisfiedBondCombinations[bondList_,maxCombos_]:=With[{bondCount=Length[bondList]},If[bondCount>0,With[{vars=Table[Unique["bonVar"],{bondCount}]},With[{result=SatisfiabilityInstances[buildBooleanExpressionForBondSelection[bondList,vars],vars,maxCombos]},Clear@@vars;result]],{}]]
In[29]:=
findSatisfiedBondCombinations[bondPositions,5]
Out[29]=
{{True,False,False,False,False,True,True,True,False,False},{True,False,False,False,False,True,True,False,False,False},{True,False,False,False,False,True,False,True,False,False},{True,False,False,False,False,True,False,False,False,False},{True,False,False,False,False,False,True,True,True,False}}
Now, we'll pick out which bonds positions satisfy the given list, giving us the bond lists we have sought.
In[30]:=
generatedBondLists=Map[Bond,Pick[bondPositions,#]]&/@findSatisfiedBondCombinations[bondPositions,1000];generatedBondLists[[1;;5]]
Out[31]=
{{Bond[{1,5}],Bond[{2,6}],Bond[{3,10}],Bond[{4,8}]},{Bond[{1,5}],Bond[{2,6}],Bond[{3,10}]},{Bond[{1,5}],Bond[{2,6}],Bond[{4,8}]},{Bond[{1,5}],Bond[{2,6}]},{Bond[{1,5}],Bond[{3,10}],Bond[{4,8}],Bond[{6,11}]}}
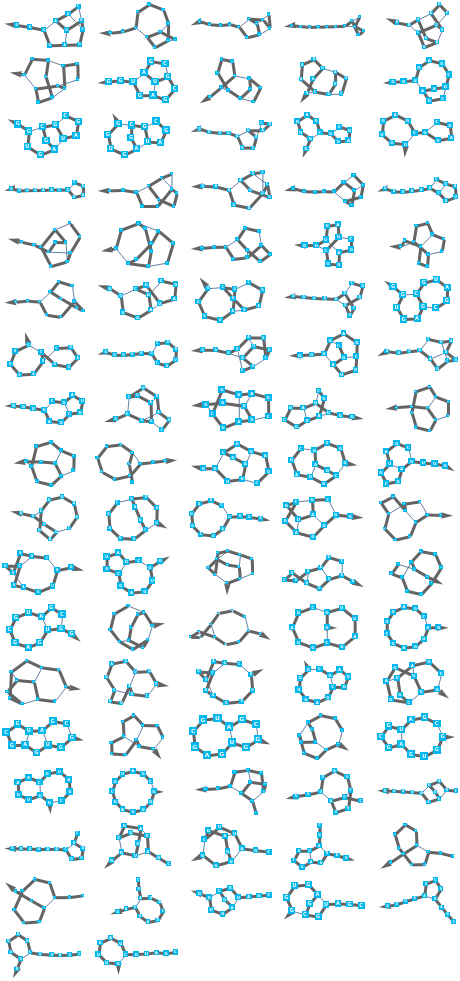
Add each of the generated bond sequences to the original bond sequence and plot them all:
In[32]:=
GraphicsGrid[Partition[BioSequencePlot/@(BioSequenceModify[seq,{"AddBond",#}]&/@generatedBondLists),UpTo[5]]]
Out[32]=